
4. Mouse proteins compared to human
Alec MacAndrew
| The draft mouse genome was published on 6th December 2002 , Waterstone et al, Nature 420, 520 - 562
Note that this is a 43 page paper (Nature averages 2 -3 pages per paper) with around 200 authors and 330 references. This is all new to science and the volume of material is more than a very fat text book if one includes the references . The detail is published not in a single paper, but in about six related papers occupying more than half of the super fat 6th December issue of Nature. |
Conservation of mouse proteins
There are two primary mechanisms that seem to have been responsible for building new mammalian proteins. The first is the organisation of well-conserved domains into new architectures; the second is duplication and co-option of new genes in gene families.
The fact that the mouse and human genome are relatively closely related allows us to study the evolution of orthologous proteins (ie proteins in different species that have diverged in the two lineages from a common ancestor.)
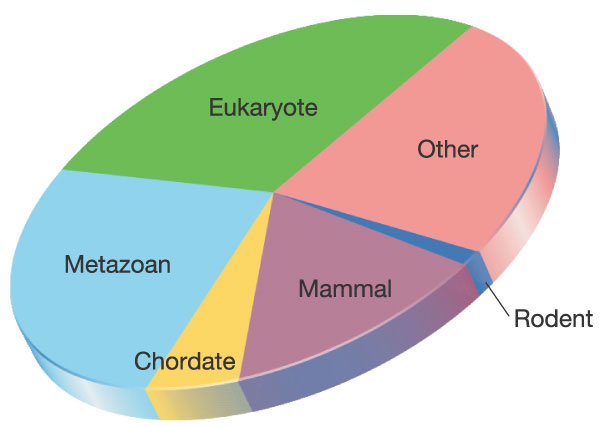
A taxonomic analysis of mouse proteins reveals that less than 1% of them are specific to mice (99% are shared with other mammals of which more than 98% are shared with man). A further 14% are specific to mammals, a further 6% are specific to chordates, a further 27% are specific to all animals, a further 29% are specific to eukaryotes and the remaining 23% are shared with all organisms including bacteria and Archaea. See Figure below:
Orthologues
There are 12,845 1:1 orthologues (genes which have descended from a common ancestor) in the mouse and human sequences. (Note that there are other orthologues in the genomes, but if there is a single duplication of a gene in either lineage, it is not a 1:1 orthologue, as it is difficult or impossible to tell which of the copies is the ancestral gene and which is the duplication).
The researchers compared complementary DNA (cDNA – that is DNA which is retrotranscribed from messenger RNA) and calculated how many amino acids in each protein were identical between mouse and man. The answer is that 70.1% of the amino acids in orthologous proteins are identical between mouse and man and the median sequence identity is 78.5%.
A further review was conducted to compare the Ka/Ks ratio. Ka is the rate of non-synonymous mutation (a non-synonymous mutation results in a different amino acid from the original code and so is subject to selective pressure in functional proteins) and Ks is the rate of synonymous mutation (a synonymous mutation results in no change of amino acid or protein and so is not subject to selective pressure even in functional proteins). In non-functional proteins this ratio = 1, as neither synonymous nor non-synonymous mutations are under selection. In most functional proteins the ratio is much less than unity as non-synonymous mutations are under strong conservative selection. In comparing mouse to man in the 12,845 pairs of orthologous proteins, we find the median Ka/Ks ratio = 0.115. Therefore most mouse and human orthologue pairs, in having a Ka/Ks ratio well below unity are shown to be under relatively strong conservative selection.
Next, the selection in regions containing known domains (ie sequences that are shorter than a gene but have functionality associated with them) was considered. It turns out that regions containing known domains have a lower Ka/Ks ratio than regions that do not contain domains, suggesting that domains are under stronger selection than the remainder of the genes. This, in effect, indicates that even within genes that are functional and code for proteins, some parts are critical and are conserved (loosely aligned with regions containing domains) and others are less critical and are less well conserved (loosely aligned with regions that do not contain domains).
Interestingly, within domains there is a hierarchy of conservation. Domains associated with nuclear functions are more conserved than domains associated with cytoplasmic functions that in turn are more conserved than domains associated with secreted portions of proteins. Secreted proteins associated with the mammalian defence and immune response systems are under less rigorous selection constraints or possibly under positive selection.
These findings can only be explained by common ancestry, mutation and selection over millions of years. There is no other explanation for the correlation of Ka/Ks ratio with regions in the genome that have function.
Further evidence for common ancestry comes SNPs (Single Nucleotide Polymorphisms) – differences in genomes that exist in different humans where the variation is a single base pair substitution and the alleles co-exist in the population. The researchers compared human alleles with mouse and found that the mouse gene is identical to the major (most common) human allele in 67% of cases (similar to the 70% amino acids which are identical between mouse and man proteins).
Gene families
Gene families are groups of related genes which have similar functionality and which have arisen by duplication of a single ancestral gene. The number of genes in the same families is different between mouse and human for some families these indicate families where developments have occurred which are specific to each lineage.
One important example is the genes associated with smelling. The different number of olfactory genes reflects the lineage-specific capability and importance of smell in the two lineages. About 20% of mouse olfactory receptor homologues are pseudogenes indicating that olfactory genes can be created by duplication and can also be silenced (and possibly revived by further mutations).
25 mouse-specific gene clusters were identified where multiple duplications had occurred to the ancestral gene (as the cluster was represented by only one or two copies in man). These gene clusters could be categorised broadly in two groups.
The first group is associated with reproduction. These gene clusters include genes which have functions which have relevance for the structure of the placenta, the survival and efficacy of spermatozoa, reproduction-related hormone metabolism and olfactory cues. These last are very interesting as they include gene clusters associated with olfactory receptor functionality as well as genes which code for pheromonal proteins such as Abpα which is a reproduction related pheromone that rodents apply to their fur by licking and which they also deposit on their surroundings. It is known that female mice prefer males with a similar Abpα which helps to isolate mouse subspecies. Additional Abpα paralogues have been found in the mouse genome, which are either once functional but now deleted pseudogenes or previously unknown but active pheromones. This very large category of mouse specific expanded gene families reflects major differences between rodent and primate lineages in fundamental reproductive features such as the structure and physiology of placenta, seminal vesicle secretion, sperm efficacy and survival and pheromones associated with mate selection and aphrodisiac functions. Thus some of the features that most distinguish mouse from human feature in expanded gene families within mouse where the families clearly result from a common ancestor preserved as a single gene in humans.
A second group of gene family clusters is associated with host defence and immunity such as major histocompatibility complex genes, anti-microbial proteins and type-A ribonucleases.
In addition two categories of gene family (Ly49 in mouse and KIR in human) are unrelated but perform similar functions in the two species and result from an instance of convergent evolution.
Interestingly, by assessing the Ka/Ks ratio in these clusters we find that they are undergoing rapid sequence evolution – much faster than the rate of evolution in other functional genes. It seems that these clusters represent a major lineage difference and they are evolving more rapidly than the rest of the functional genome. So the gene clusters associated with the strongest lineage specific differences between mouse and man are also under the weakest purifying selection - just as evolutionary theory would predict.